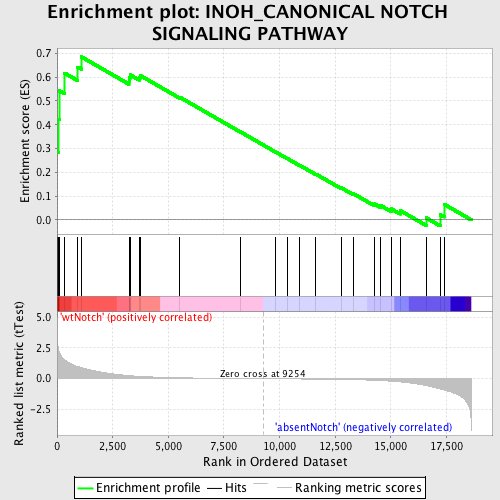
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | wtNotch |
| GeneSet | INOH_CANONICAL NOTCH SIGNALING PATHWAY |
| Enrichment Score (ES) | 0.68542403 |
| Normalized Enrichment Score (NES) | 1.624165 |
| Nominal p-value | 0.0062111802 |
| FDR q-value | 0.17734835 |
| FWER p-Value | 0.87 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NOTCH1 | 3390114 | 0 | 4.962 | 0.2840 | Yes | ||
| 2 | NOTCH3 | 3060603 | 51 | 2.482 | 0.4234 | Yes | ||
| 3 | DTX1 | 5900372 | 95 | 2.127 | 0.5429 | Yes | ||
| 4 | DTX3 | 1690458 | 340 | 1.524 | 0.6170 | Yes | ||
| 5 | PSEN1 | 130403 2030647 6100603 | 922 | 0.996 | 0.6427 | Yes | ||
| 6 | DTX2 | 4210041 7000008 | 1091 | 0.904 | 0.6854 | Yes | ||
| 7 | ITCH | 1410731 5700022 | 3232 | 0.248 | 0.5845 | No | ||
| 8 | NOTCH4 | 2450040 6370707 | 3248 | 0.246 | 0.5978 | No | ||
| 9 | RFNG | 6290086 | 3279 | 0.242 | 0.6100 | No | ||
| 10 | FBXW7 | 4210338 7050280 | 3691 | 0.190 | 0.5987 | No | ||
| 11 | PSEN2 | 130382 | 3736 | 0.186 | 0.6070 | No | ||
| 12 | NOTCH2 | 2570397 | 5506 | 0.074 | 0.5161 | No | ||
| 13 | DLL4 | 6400403 | 8253 | 0.014 | 0.3692 | No | ||
| 14 | DLL3 | 4010093 | 9826 | -0.008 | 0.2851 | No | ||
| 15 | ADAM10 | 3780156 | 10357 | -0.015 | 0.2575 | No | ||
| 16 | JAG1 | 3440390 | 10896 | -0.024 | 0.2299 | No | ||
| 17 | JAG2 | 1500341 | 11635 | -0.036 | 0.1923 | No | ||
| 18 | MFNG | 1770242 4280348 | 12767 | -0.063 | 0.1350 | No | ||
| 19 | NUMB | 2450735 3800253 6350040 | 13320 | -0.081 | 0.1100 | No | ||
| 20 | RBPJ | 2360164 5550136 5720402 | 14258 | -0.130 | 0.0670 | No | ||
| 21 | DLL1 | 1770377 | 14531 | -0.150 | 0.0610 | No | ||
| 22 | FURIN | 4120168 | 15017 | -0.199 | 0.0463 | No | ||
| 23 | ADAM17 | 360292 | 15422 | -0.257 | 0.0393 | No | ||
| 24 | LFNG | 5360711 | 16588 | -0.557 | 0.0085 | No | ||
| 25 | CREBBP | 5690035 7040050 | 17223 | -0.828 | 0.0218 | No | ||
| 26 | MAML1 | 2760008 | 17404 | -0.928 | 0.0652 | No |

